occular correction
EMEGS (Qt version) offers an occular correction procedure based on hierarchichal mutiple linear regression: the vertical (VEOG), the horizontal (HEOG) and the radial electrooculogram (REOG) are used as independant variables in a multiple linear regression for each EEG channel. The corresponding beta weights (slopes) are tested for statistical significance and non-significant channels are removed from the model until only significant channels remain.
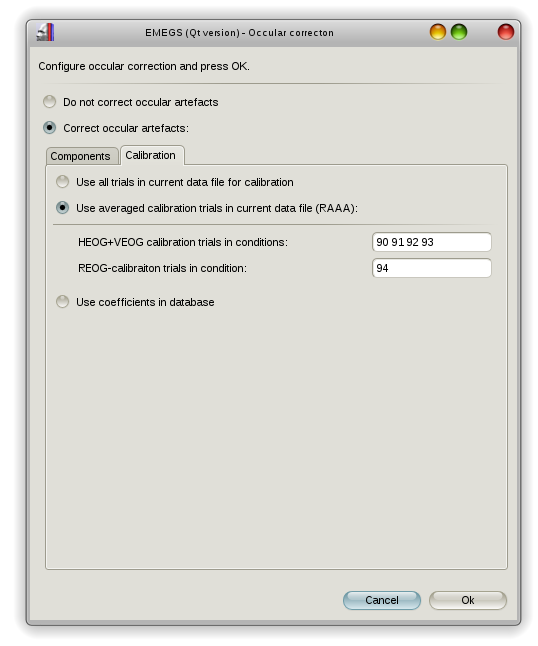
You can either use all your experimental data to calulate the beta weights or use the revised artefact-aligned-average procedure (RAAA) as suggested by Croft & Barry (2000). This procedure requires data from a specific calibration sequence and estimates VEOG/HEOG and REOG betas separately, from saccade and blink data respectively.
EMEGS (Qt version) will prompt you to configure the occular correction when you press the “Resume analysis”-button after the data segmentation step. Here you can select the sensors for VEOG, HEOG and REOG. During previous steps, EMEGS (Qt versions) usually creates an individual sensor configuration file next to the data file (to account for instance for dropped channels when processing BDF files, that can contain non-EEG channels). This file is selected by default and an @ -sign is put in front of the filename to indicate, that ist is a dynamically created individual file.
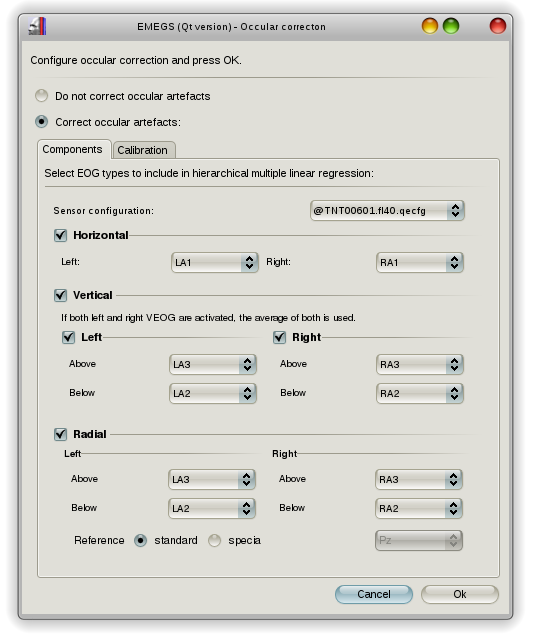
Sensor configuration can contain information for occular corrections, namely the keywords HEOG_L, HEOG_R, LVEOG_B, LVEOG_T, RVEOG_B, RVEOG_T and REOGRef in the “Occular” column, indicating the left HEOG sensor, right HEOG sensor, left-bottom VEOG sensor, left-top VEOG sensor, right-bottom VEOG sensor, right-top VEOG sensor and REOG reference sensor respectively. These channels will be selected automatically, if a sensor configuration file (or an adopted version of it) is selected. An example of a configuration file is shown below:
Type Name InRawFile RefWeight Occular Theta Phi Rho
EEG Fp1 Yes 0 No -1.6057 -1.25664 0.09
EEG AF7 Yes 0 No -1.6057 -0.942478 0.09
EEG AF3 Yes 0 No -1.29154 -1.13446 0.09
EEG F1 Yes 0 No -0.872665 -1.18682 0.09
… … … … … … … …
EEG LA1 Yes 0 HEOG_L -1.96175 -0.842842 0.09
EEG LA2 Yes 0 LVEOG_B 2.17302 1.9583 0.09
EEG LA3 Yes 0 LVEOG_T 1.77549 1.8746 0.09
EEG RA1 Yes 0 HEOG_R 1.96175 0.842842 0.09
EEG RA2 Yes 0 RVEOG_B 2.17302 1.1833 0.09
EEG RA3 Yes 0 RVEOG_T 1.77549 1.26698 0.09
EEG A1 Yes 0.5 No -2.00713 0.1 0.09
EEG B1 Yes 0.5 No 2.00713 -0.1 0.09
STATUS Status Yes 0 No NAN NAN NAN
On the second page of the Occular-correction-dialog, you can select the beta calibration procedure, either using all extracted, or using averaged calibration trials as suggested by Croft and Barry (2000), or loading betas from the database file in the study folder (not yet supported). RAAA-calibration trials for HEOG and VEOG should be 4 trial types: movement to the left, movement to the right, movement up, movement down, REOG calibration trials should be blinks.
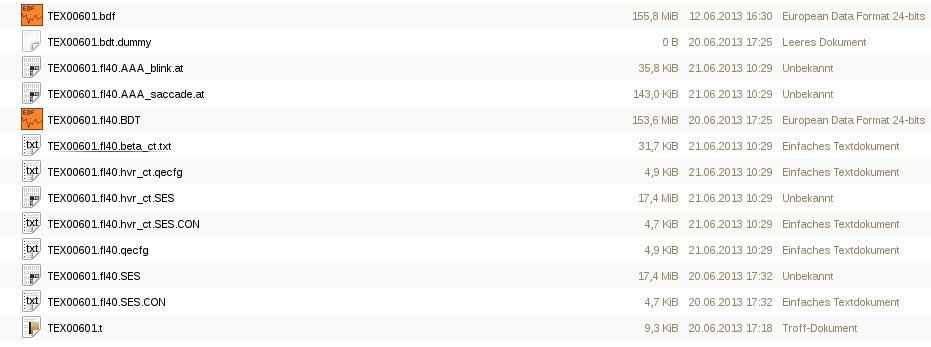
After the occular correction, the corrected trials are displayed, and a couple of new files have appeared in your data folder, most importantly the corrected SES-file, the name of which is constructed depending on your occular correction configuration ** (hvr for HEOG, VEOG and REOG, ** _ct for calibrated with calibration trials, or _at for calibrated with all trials). A text-file containing the regression betas is also created, which looks like this:
…
Sensor P3:
Component Beta Std.dev. T P Step
Intercept -9.66859498e-12 1.64139629e-01 -5.89046962e-11 1.0000000 2
VEOG 1.17232219e-01 3.85906319e-03 3.03784139e+01 0.0000000* 2
HEOG 1.66331788e-03 1.29132049e-03 1.28807518e+00 0.1987550 1
REOG -3.53498120e-01 3.63414858e-03 -9.72712348e+01 0.0000000* 1
…
If the RAAA-procedure was used, two additional files are created (*AAA_saccade.at & *AAA_blink.at) containing the artefact-aligned averages.
Croft, R.J., Barry, R.J. (2000). Removal of ocular artifact from the EEG: a review. Clinical neurophysiology, 30(1), 5-19.
