welcome
Created by: Root, Last modification: Sat 18 of Jul, 2020 (05:04 UTC) by emegsadmin
Welcome to emegs.org!
EMEGS (ElectroMagnetic EncaphaloGraphy Software) is an open source software for psychophysiological data analysis. There are two versions available: the original MATLAB based package which requires the MATLAB environment and a stand-alone Qt based version.
To download EMEGS, register/login first.
Numerous studies have used EMEGS for preprocessing, visualization or statistical analysis. You can find a list of references here.
The current version of EMEGS for Matlab is 3.1, the developer version is 3.2.
See the "What's new"-page for new features in EMEGS for Matlab.
The current version of EMEGS (Qt version) is 2.0, the developer version is 2.1.
See the "What's new"-page for new features in EMEGS (Qt version).
For support in using EMEGS please refer to the email discussion list and the email archive at https://lists.sourceforge.net/lists/listinfo/emegs-user.
Please refer to EMEGS (all versions) in publications as follows:
Peyk, P., De Cesarei, A., & Junghöfer, M. (2011). Electro Magneto Encephalograhy Software: overview and integration with other EEG/MEG toolboxes. Computational Intelligence and Neuroscience, Volume 2011, Article ID 861705.
You can also visit our project for news on Researchgate.
EMEGS now offers support for a range of permutation based cluster mass tests for coefficients from t-tests, correlations, custom contrasts and also full factorial ANOVA. The later is realized through residualization of the data for lower order effects. A serial rejection algorithm is available to reduce masking effects. All features are configured on a GUI, no coding is necessary.
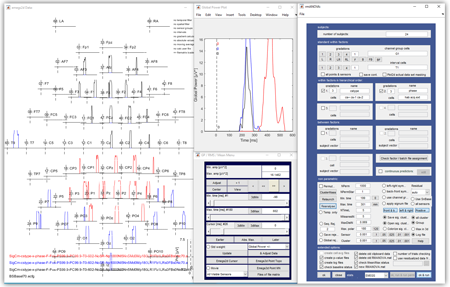
EMEGS (ElectroMagnetic EncaphaloGraphy Software) is an open source software for psychophysiological data analysis. There are two versions available: the original MATLAB based package which requires the MATLAB environment and a stand-alone Qt based version.
To download EMEGS, register/login first.
Numerous studies have used EMEGS for preprocessing, visualization or statistical analysis. You can find a list of references here.
The current version of EMEGS for Matlab is 3.1, the developer version is 3.2.
See the "What's new"-page for new features in EMEGS for Matlab.
The current version of EMEGS (Qt version) is 2.0, the developer version is 2.1.
See the "What's new"-page for new features in EMEGS (Qt version).
For support in using EMEGS please refer to the email discussion list and the email archive at https://lists.sourceforge.net/lists/listinfo/emegs-user.
Please refer to EMEGS (all versions) in publications as follows:
Peyk, P., De Cesarei, A., & Junghöfer, M. (2011). Electro Magneto Encephalograhy Software: overview and integration with other EEG/MEG toolboxes. Computational Intelligence and Neuroscience, Volume 2011, Article ID 861705.
You can also visit our project for news on Researchgate.
News: support for permutation-based cluster mass tests
EMEGS now offers support for a range of permutation based cluster mass tests for coefficients from t-tests, correlations, custom contrasts and also full factorial ANOVA. The later is realized through residualization of the data for lower order effects. A serial rejection algorithm is available to reduce masking effects. All features are configured on a GUI, no coding is necessary.


 home
home